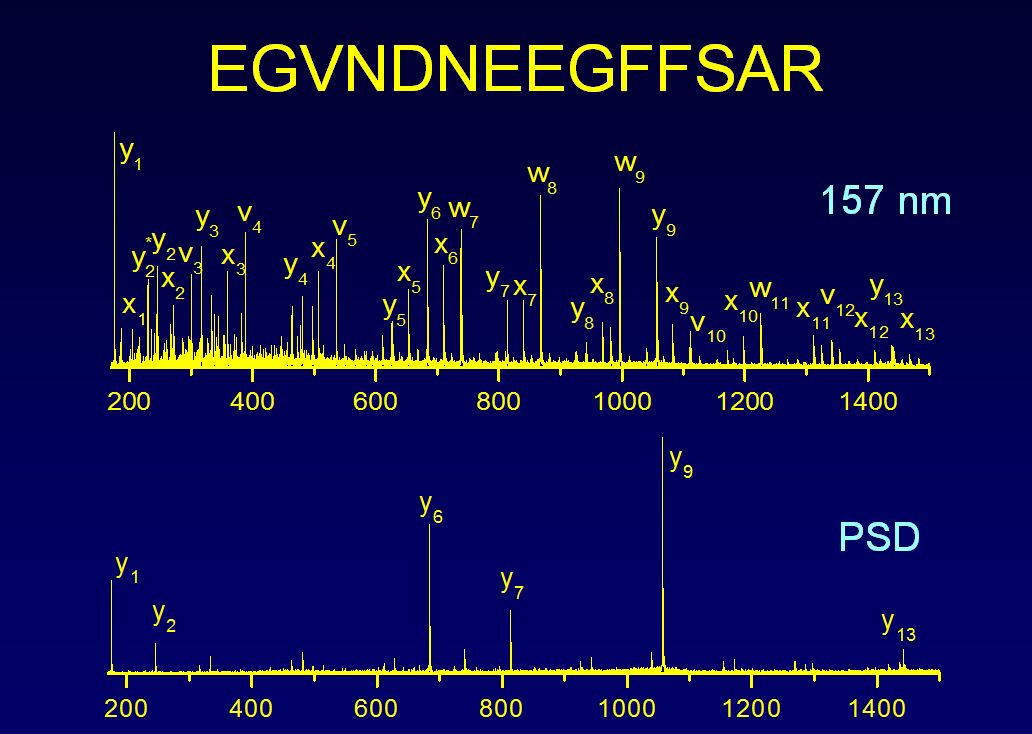
Detailed sequence and structural information about peptides and proteins is sought to improve our understanding of cellular processes and to develop novel therapeutics. Mass spectrometry of biological molecules enables us to analyze cellular constituents. Laser induced photofragmentation of peptide ions can provide a number of technological advantages over collision induced dissociation that include increased sequence coverage and production of a wide variety of fragment ion types. We perform our photofragmentation studies on a number of instrumental platforms.
Computer algorithms commonly employed to identify proteins compare collisional fragmentation spectra of peptide ions with predictions derived from protein sequence databases. Agreement is often marginal. Photofragmentation of peptide ions with vacuum ultraviolet laser light yields richer fragmentation patterns that can be directly interpretable. Photofragmentation data are being analyzed with a combination of de novo sequencing and database matching to improve the reliability of identifications and our fundamental understanding of the phenomenon.
An antibody is a large protein whose unique structure enables it to specifically bind target antigens. The structure consists of two heavy and two light chains that contain various disulfide bonds that stabilize the tertiary structure. The variable part of the Fab region is of particular interest. It contains small sequences called complementarity determining regions (CDRs) that are the main binding sites for antigens. There are three CDRs in each heavy chain and three in each light chain. Antibodies are not encoded by the genome of the host organism, so being able to sequence antibody proteins can generate new biological information. A high-throughput method for digesting and de novo sequencing these large proteins may enable the development of antibodies that bind antibodies more tightly and more selectively.
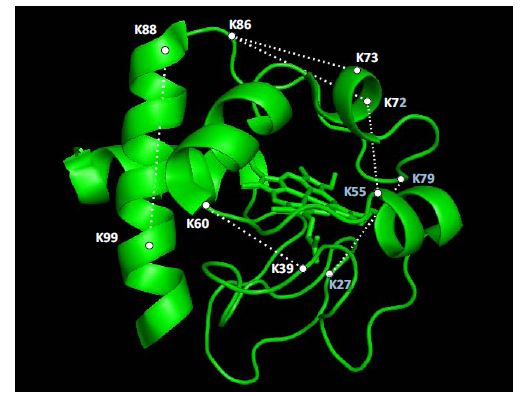
The structures of most proteins that are bound to surfaces or involved in interactions with other molecules cannot be determined by x-ray crystallography. The combination of chemical labeling and cross-linking can provide key structural information that allows the determination of regions of interaction. Liquid chromatography/mass spectrometry methods are being used to analyze interacting proteins. We are currently identifying and locating ribosome-associated proteins that participate in the protein translation process or that modify nascent proteins soon after they are synthesized.
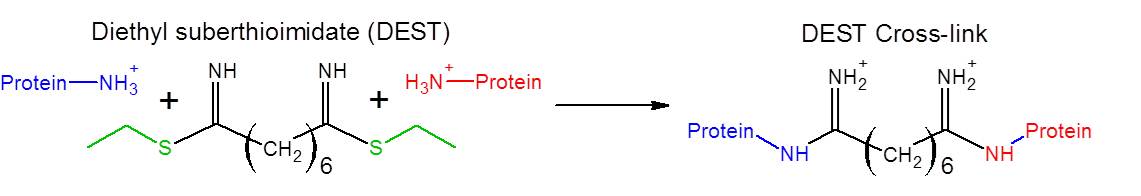
Methods utilizing novel thioimidate reagents have recently been developed to aid in the understanding of protein structure. Below is a scheme of the reaction of DEST (a novel cross-linker recently developed in the group) with a protein.
References:
M.A. Lauber and J.P. Reilly, Novel Amidinating Cross-Linker for Facilitating Analyses of Protein Structures and Interactions. Analytical Chemistry 2010, 82 (18), 7736-7743.M.A. Lauber and J.P. Reilly, “Use of Homobifunctional Thioimidates for Protein Cross-linking and Analyte Enrichment,” patent pending (2010).
Janecki, D. J.; Beardsley, R. L.; Reilly, J. P., Probing Protein Tertiary Structure with Amidination. Analytical Chemistry 2005, 77 (22), 7274-7281.
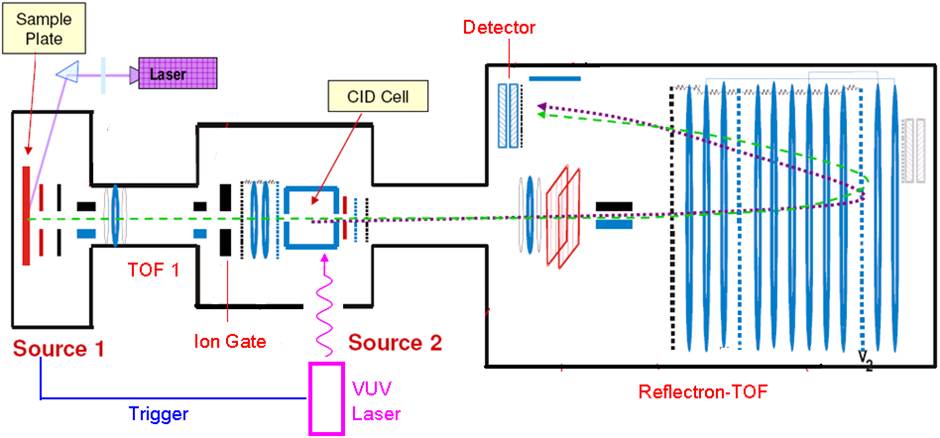
We are currently developing an improved photodissociation TOF-TOF mass spectrometer. Interfaces between the mass spectrometer and a 157nm VUV laser are being fabricated that enable light to be transmitted through vacuum. Instrumental improvements are being incorporated in order to obtain higher-quality photodissociation data that will improve our capabilities for de novo sequencing.
| © 2014 Reilly Group. All rights reserved. |